A recent study has collected phenotypic data of chickpea (Cicer arietinum L.) which can now be linked with the genotypic data of these lines. This will enable genome-wide association mapping with the aim of identifying loci that underlie salinity tolerance – an important step in developing salt tolerant chickpeas.
In this study, Judith Atieno and co-authors utilised image-based phenotyping at the Australian Plant Phenomics Facility here at Waite to study genetic variation in chickpea for salinity tolerance in 245 diverse accessions (a diversity collection, known as the Chickpea Reference Set).
Chickpea is an important legume crop, used as a highly nutritious food source and grown in rotation with cereal crops to fix nitrogen in the soil or to act as a disease break. However, despite its sensitivity to salt, chickpea is generally grown in semi-arid regions which can be prone to soil salinity. This results in an estimated global annual chickpea yield loss of between 8–10%.
The study found, on average, salinity reduced plant growth rate (obtained from tracking leaf expansion through time) by 20%, plant height by 15% and shoot biomass by 28%. Additionally, salinity induced pod abortion and inhibited pod filling, which consequently reduced seed number and seed yield by 16% and 32%, respectively. Importantly, moderate to strong correlation was observed for different traits measured between glasshouse and two field sites indicating that the glasshouse assays are relevant to field performance.
Using image-based phenotyping, we measured plant growth rate under salinity and subsequently elucidated the role of shoot ion independent stress (resulting from hydraulic resistance and osmotic stress) in chickpea.
Broad genetic variation for salinity tolerance was observed in the diversity panel with seed number being the major determinant for salinity tolerance measured as yield. The study proposes seed number as a selection trait in breeding salt tolerant chickpea cultivars.
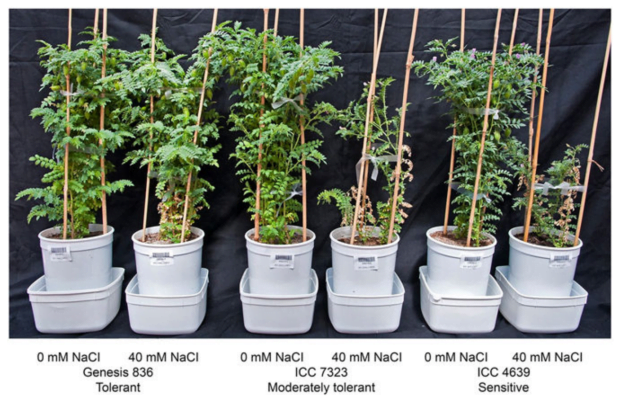
Genotypic variation for salinity tolerance in the Chickpea Reference Set. Varying levels of salinity tolerance exhibited by different chickpea genotypes. Exposure of sensitive genotypes to 40 mM NaCl caused severe stunted growth, leaf damage, and led to less number of reproductive sites (flowers and pods) compared to moderately tolerant and tolerant genotypes.
The rapid development of new, high-resolution and high-throughput phenotyping technologies in plant science has provided the opportunity to more deeply explore genetic variation for salinity tolerance in crop species and identify traits that are potentially novel and relevant to yield improvement. The Australian Plant Phenomics Facility provides state-of-the-art phenotyping and analytical tools and expertise in controlled environments and in the field to help academic and commercial plant scientists understand and relate the performance of plants to their genetic make-up. A dedicated cross-disciplinary team of experts provides consultation on project design and high quality support.
To read the full paper in Scientific Reports, “Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping” (doi:10.1038/s41598-017-01211-7), click here.
To find out more about the Australian Plant Phenomics Facility and how we can support your research click here.
Republished from the Australian Plant Phenomics Facility blog.
